

| Ligand-Protein Complex | Affinity | Interacting Residue, the amino acid and its position | Hydrogen Bond Length | Interpretation | |
|---|---|---|---|---|---|
| 1 | 1XJD & STU | KPCT_HUMAN, Affinity=0.00033µM | [ASP]508:A, [LEU]461:A | 2.65 Å, 2.58 Å | Binding: Very strong Short hydrogen bond lengths coupled with an extremely low affinity value indicate very strong binding. (negative charge, ASP) |
| 2 | 2Z7R & STU | KS6A1_HUMAN, Affinity=0.003µM | [ASP]148:A, [LEU]144:A | 3.09 Å, 2.52 Å | Binding: StrongA mix of bond lengths, but a very low affinity suggests strong overall binding. (negative charge, ASP) |
| 3 | 2OIC & STU | IRAK4_HUMAN, Affinity=0.004µM | [MET]265:A | 2.77 Å | Binding: Moderately StrongA single moderately strong bond, contributes to moderately strong binding. |
| 4 | 1OKY & STU | PDPK1_HUMAN, Affinity=0.0065µM | [ALA]162:A, [GLU]166:A, [GLU]209:A | 3 Å, 2.45 Å, 3.09 Å | Binding: Moderately StrongMultiple bonds with one notably short bond, indicate moderately strong binding. (negative charge, GLU & GLU) |
| 5 | 1NVR & STU | CHK1_HUMAN, Affinity=0.0078µM | [CYS]87:A, [GLU]134:A, [GLU]91:A | 2.76 Å, 2.92 Å, 2.69 Å | Binding: Moderatemultiple bonds of moderate lengths suggest moderate binding strength. (negative charge, GLU & GLU) |
| 6 | 1YHS & STU | PIM1_HUMAN, Affinity=0.01µM | [ASP]128:A, [GLU]171:A | 2.81 Å, 2.9 Å | Binding: ModerateModerately strong hydrogen bonds reflect moderate affinity. (negative charge, ASP & GLU) |
| 7 | 1SM2 & STU | ITK_HUMAN, Affinity=0.01µM | [ARG]486:A, [MET]438:A | 2.55 Å, 2.64 Å | Binding: ModerateRelatively short bond lengths contribute to moderate binding. (positive charge, ARG) |
| 8 | 1XBC & STU | KSYK_HUMAN, Affinity=0.012µM | [ALA]451:A, [ARG]498:A | 2.84 Å, 3.08 Å | Binding: ModerateA mix of bond lengths, suggesting moderate binding strength. (positive charge, ARG) |
| 9 | 1Q3D & STU | GSK3B_HUMAN, Affinity=0.015µM | [VAL]135:A | 2.96 Å | Binding: Slightly WeakA single bond of moderate length and higher affinity indicates slightly weaker binding. (Only one bond) |
| 10 | 1U59 & STU | ZAP70_HUMAN, Affinity=0.0558µM | [ALA]417:A, [ARG]465:A | 2.63 Å, 2.89 Å | Binding: WeakModerate bond lengths, but higher affinity suggests weaker binding. (positive charge, ARG) |
| 11 | 3HMO & STU | TTK_HUMAN, Affinity=0.102µM | [GLY]605:A | 2.61 Å | Binding: WeakestDespite a relatively short bond, the high affinity number suggeste the weakest binding. (only one hydrogen bond) |
Correlation of Bond Length and Affinity: There seems to be a general correlation where lower affinity values (stronger binding) are often accompanied by shorter hydrogen bond lengths. However, this is not a strict rule, as other factors also influence binding strength. Hydrogen bond lengths are a good indicator of bond strength, but they are not the only factor. The overall fit of the ligand in the protein's active site and other types of interactions (like hydrophobic interactions) also play a role.
Strongest Binding: The strongest binding is observed in 1XJD & STU (KPCT_HUMAN) and 2Z7R & STU (KS6A1_HUMAN), indicated by very low affinity values and short hydrogen bond lengths.
Weakest Binding: The weakest binding is noted in 3HMO & STU (TTK_HUMAN), as suggested by the highest affinity value among the entries, despite having a relatively short hydrogen bond.
Multiple Bonds vs. Single Bonds: Complexes with multiple hydrogen bonds often exhibit stronger binding, indicating a cumulative effect of multiple interactions.
Role of Specific Residues: The nature of the interacting amino acids also plays a role in the binding strength. Certain residues might form stronger or more specific interactions with staurosporine.
In biochemistry and pharmacology, affinity refers to the strength of the interaction between a drug (like staurosporine) and its target (like a protein kinase). It is a measure of how well a drug can bind to a particular target. When a compound has a "high affinity" for a target, it means that it binds very strongly to that target. This is reflected in low numerical values in affinity measurements. For example, an affinity of 0.00033 µM (micromolar) is considered very high because the drug can bind effectively even at this very low concentration. Conversely, a higher numerical value in affinity measurements (like 0.102 µM) indicates that a higher concentration of the drug is needed to effectively bind to the target, which in turn means the binding is weaker.
This relationship of binding affinity is calculated by the dissociation constant (Kd). The constant is inversely related to the strength of the binding - the smaller the Kd, the stronger the binding, and vice versa. Therefore, in the results above a higher numerical affinity value indicates that the drug binds less tightly to the protein, characterising this as a weaker interaction. Kd is defined as the concentration of the ligand at which half of the available binding sites on the protein are occupied. In other words, it is the point at which the concentration of the bound ligand-protein complex is equal to the concentration of the unbound ligand and protein (Malvern Panalytical, n.d.).
The relationship can be represented by the equation:
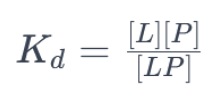 where:
[L][L] is the concentration of the free ligand.
[P][P] is the concentration of the free protein.
[LP][LP] is the concentration of the ligand-protein complex.
Low Kd Value: A low Kd indicates that the ligand binds very tightly to the protein. This is because even at low ligand concentrations, a significant portion of the protein is in the bound state. In practical terms, a drug with a low Kd value has a high affinity for its target, meaning it is very effective at binding even at low concentrations.
High Kd Value: Conversely, a high Kd value suggests that higher concentrations of the ligand are needed to significantly occupy the binding sites on the protein. This translates to a lower affinity of the ligand for the protein (Lieberman et al., 2013).
Application in Drug Design
In pharmacology, Kd is crucial for drug design and understanding drug-receptor interactions. A drug with a lower Kd value for its target is typically more desirable, as it means the drug can exert its effects at lower concentrations, potentially reducing the risk of side effects and increasing its efficacy.
where:
[L][L] is the concentration of the free ligand.
[P][P] is the concentration of the free protein.
[LP][LP] is the concentration of the ligand-protein complex.
Low Kd Value: A low Kd indicates that the ligand binds very tightly to the protein. This is because even at low ligand concentrations, a significant portion of the protein is in the bound state. In practical terms, a drug with a low Kd value has a high affinity for its target, meaning it is very effective at binding even at low concentrations.
High Kd Value: Conversely, a high Kd value suggests that higher concentrations of the ligand are needed to significantly occupy the binding sites on the protein. This translates to a lower affinity of the ligand for the protein (Lieberman et al., 2013).
Application in Drug Design
In pharmacology, Kd is crucial for drug design and understanding drug-receptor interactions. A drug with a lower Kd value for its target is typically more desirable, as it means the drug can exert its effects at lower concentrations, potentially reducing the risk of side effects and increasing its efficacy.
Malvern Panalytical. (n.d.). Binding affinity. Binding Affinity. Accessed 13 Dec. 2023 from https://www.malvernpanalytical.com/en/products/measurement-type/binding-affinity Lieberman, M., Peet, A., & Marks, D. B. (2013). Marks’ Basic Medical Biochemistry: A Clinical Approach (4th ed., p. 97). Wolters Kluwer.